KDDart Data Access Layer

This introduction provides programmers with an overview of the Data Access Layer (DAL), the RESTful Application Programming Interface (API) of the KDDart Knowledge Discovery System.
Introduction
Objective
This introduction provides programmers with an overview of the Data Access Layer (DAL), the RESTful Application Programming Interface (API) of the KDDart Knowledge Discovery System.
Along with other supporting information this document will assist a programmer either to develop their own applications or enhance existing applications to access KDDart.
Upon conclusion of this document you should:
- Have a basic understanding of the DAL RESTful API;
- Have seen some basic code examples;
- Know where to find further help documentation for DAL; and
- Be better prepared to start using and implementing DAL in your program(s).
Audience
This document is intended for the following audience:
| Role | Responsibility |
| Programmer | Programmers who are new to using DAL to build or maintain systems using KDDart. |
| Technical User | Technical users who want to get involved at a deeper, coding level. |
Overview
The Data Access Layer (DAL) plays a major role within the KDDart environment. From its inception it was designed to deliver RESTful API architected services to applications that needed data to be fed into or consumed from the KDDart database layer.
The KDDart environment consists of a three layer architecture which is illustrated in the following diagram:
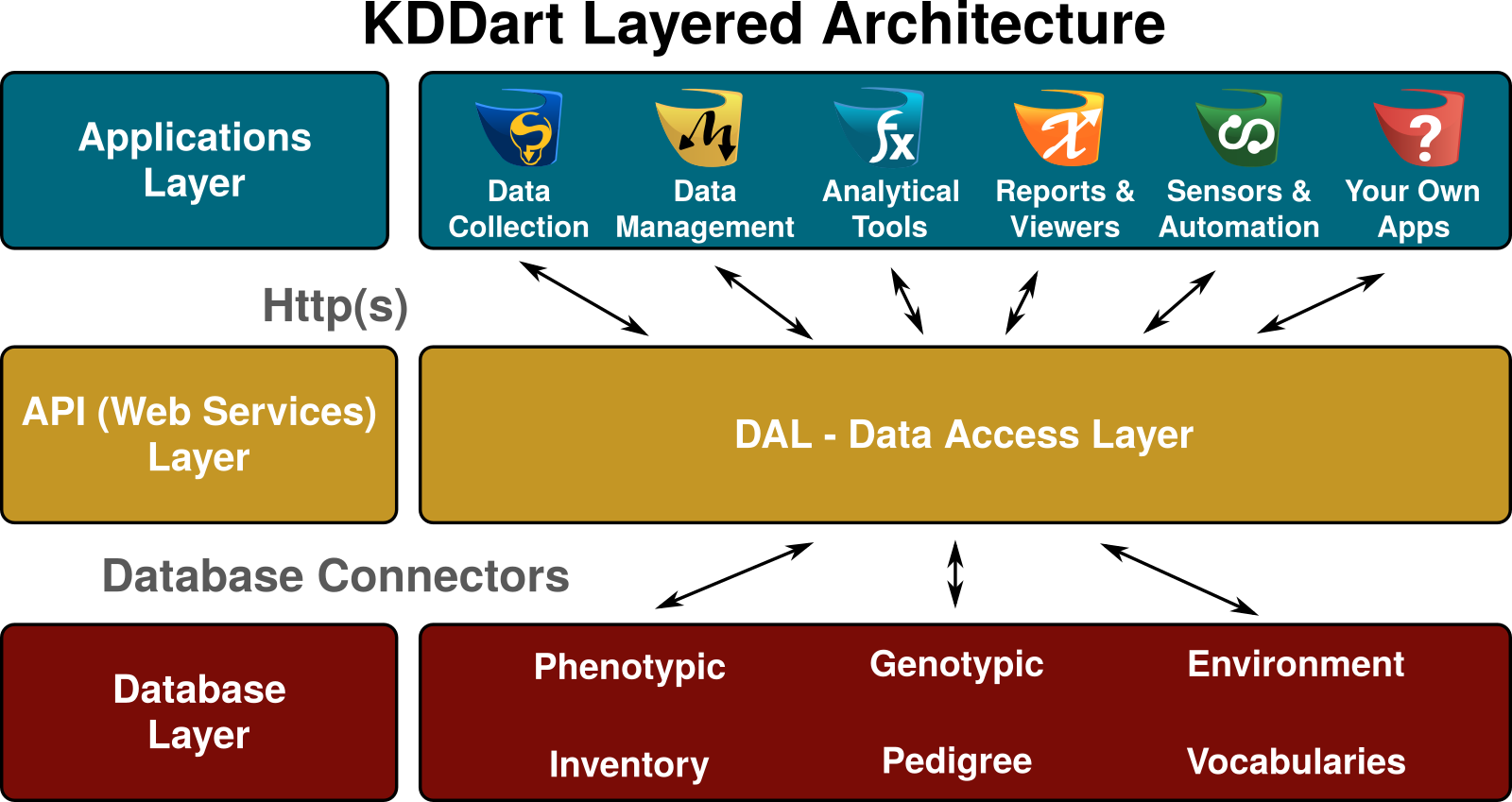
KDDart was developed to offer a flexible storage solution, to cater for a broad spectrum of data types in a domain requiring increasing data storage volumes. Whilst the database can be configured to different client requirements, having static, hard coded software data access would not efficiently accomodate such ongoing changes. Separation of the data access services, by creating a web enabled RESTful API, ensures the inevitable configuration changes are handled much more efficiently whilst providing stability and functionality for existing data repositories. Use of an API approach also offers much more opportunity to integrate KDDart services into other existing third party applications.
Designed from the onset to include a Data Access Layer using current web technology (ie a RESTful API) the system is able to support a secure, more efficient and a single, standardised gateway for applications to use.
For the developer, their software can be more robust and predictable by consuming predefined services that manage complex data dependencies for them. They don't need to be an expert with the data.
For the user, they can utilise applications which store their raw data in a central repository knowing the integrity of that data is managed by a robust Data Access Layer.
Key features of the KDDart system include:
- All access to KDDart is through the DAL web service interface;
- A sophisticated and comprehensive tool set, accessible through the DAL web interface;
- DAL is designed to ensure data integrity travelling from client to database;
- Graphical User Interface (GUI) applications also be developed to use DAL;
- It can be a Software as a Service (!SaaS) over the Internet;
- Data exchange support for XML, CSV and JSON (with !GeoJSON); and
- DAL has no visual user interface (GUI or otherwise).
Programmers do need to have an understanding of DAL to be able to efficiently develop KDDart applications such as:
- Analytical tools;
- Graphical User Interface applications; or
- Automatic data logger robots.
Functionality
The DAL API provides an extensive range of functionality for the programmer. The following table illustrates the key functional groups that are available
| Functional Groups | Description |
|---|---|
| System | Reference for the DAL system calls which provides core system administration and authentication functionality. Designed for a system management interface although some functions are accessible to all users in all parts of the system. |
| Configuration | API to configure KDDart system settings and general administration of various system components required to manage the system's back-end. |
| Vocabularies | Vocabulary definitions of various ontologies in the KDDart. These include Traits, Treatments, Breeding Methods and other controlled vocabulary definitions as required by the program or industry. |
| Germplasm | API for KDDart germplasm which supports a multi genus database in which genotypes and specimens form the core of the system along with it's ability to manage pedigree information. |
| Experiments | API for trials and experiments performed on a group of specimens or genotypes. Each specimen/genotype is grown in a trial unit (often referred to as 'plot' for cereal crops).A Trial Unit is the smallest unit for which trait/phenotypic data is collected. |
| Inventories | Seed, samples and DNA inventories that support management of seed and/or plant material which may be destined for long term storage. |
| Markers and DNA | API for KDDart Markers and DNA provides functionality for the management of DNA profiles (markers) and the integration of this information with specimens (grown in trials).Module has the flexibility to cater for various types of markers and other DNA information. |
| Environment | API for environment and GIS data which provides for the recording of information for a variety of measurable environmental conditions as well as images such as aerial photographs. |
| Other | Other useful features such as multimedia that extend the functionality of the API, including programmatic help and utility functions. |
Data Integrity
Large data volumes coupled with many complex relationships is what KDDart is designed to efficiently store. The DAL is the custodian of data integrity and it is in this role that it may be considered to be the sentry, being the single point of entry and exit for all data in KDDart.
Not to be confused with data security, data integrity is about ensuring client information is stored accurately. The DAL ensures data captured at a computer or a device is identical to the data received at the server for adding or updating in KDDart. Regardless of whether this data is moving over the internet or on an internal network, the DAL checks to ensure it hasn’t changed, either deliberately or accidently, whilst in transit to the database.
The integrity of the data is assured through design, however the confidentiality of that information is at the discretion of the client's design requirements and hence the configuration of their system. Confidentiality assurance is provided through using HTTPS in preference to HTTP as the communication protocol when required.
Whilst referring to security it is worth noting, that even though a site’s requirement is for low confidentiality, no authentication criteria such as passwords are sent ‘in the clear’.
DAL Data Management Operations
This section introduces some of the DAL’s operations, however for simplicity several important factors have been set aside and are not detailed, namely authentication (i.e. login) and group selection. Prior to any operations in the system the user must have already successfully authenticated and have the appropriate permission to perform the operations on the data.
Note: The full URLs are not displayed in the examples that follow which are installation dependent. DAL URLs follow the REST convention so the URL to list 50 records in the genotype table is:
https://example.diversityarrays.com/dal/list/genotype/50/page/1
Two of the main DAL data management operations, used across all entities, which are:
- Read;
- Add; and
- Update.
Read (List and Get) Operations
For the KDDart read operation a client can either list records for an entity or get an individual record. In either case, the steps to create these operations follow the simple URL formation as follows:
List Syntax:
list/<table name>[/<number of records per page>/page/<page number>]
Get Syntax:
get/<table name>/<record id>
The parameters for these operations are described in the following table:
| Parameter | Description |
|---|---|
| Table Name | The table or entity name in KDDart. For example, these could be genotype, specimen, trait, treatment, trial, etc. |
| number of records per page | For tables where pagination is available this value indicates the number of records to retrieve for a ‘page’ |
| /page/ | The page attribute |
| page number | The number of the page to return |
| Record id | The individual record number to retrieve – for the Get operation |
Using the genotype table as an example, the URL to list 50 genotype records from KDDart would be:
list/genotype/50/page/1.
To list all the genotypes in the table use:
list/genotype
The URL to retrieve or get an individual genotype record with record id=5 would be:
get/genotype/5
The following example illustrates the returned XML data for one genotype table record. It is not intended for further dissection, merely to illustrate how information can be returned to the program.
<DATA>
<Pagination Page="1" NumOfRecords="695" NumOfPages="139" NumPerPage="1"/>
<RecordMeta TagName="Genotype"/>
<Genotype AccessGroupPerm="5"
AccessGroupId="0"
GenotypeName="Geno_5327164"
GenotypeId="695"
AccessGroupPermission="Read/Link"
OtherPermission="None"
addAlias="genotype/695/add/alias"
chgPerm="genotype/695/change/permission"
OtherPerm="0"
OwnGroupPerm="7"
CanPublishGenotype="0"
OriginId="0"
GenotypeNote="none"
SpeciesName="Testing"
GenotypeColor="black"
OwnGroupPermission="Read/Write/Link"
OwnGroupName="admin"
GenusName="Genus_6428101"
GenusId="10"
AccessGroupName="admin"
GenotypeAcronym="T"
chgOwner="genotype/695/change/owner"
UltimatePermission="Read/Write/Link"
delete="delete/genotype/695"
OwnGroupId="0"
update="update/genotype/695"
UltimatePerm="7"/>
</DATA>
A positive ‘feature’ of the RESTful interface approach is a developer, or advanced user, can perform some of these operations from a browser to quickly check for the type of results being returned. It is also useful for checking if they are using the correct syntax for the URL under construction.
In the previous examples, replacing ‘genotype’ with ‘specimen’ in the URL
list/specimen/50/page/1
would retrieve 50 specimen names from KDDart.
Alternatively to retrieve an individual specimen record with a record id=11 the URL would be:
get/specimen /11
In addition to what has just been described, illustrating pagination, further advanced list operations are available, which cater for:
- Field selection;
- Filtering; and
- Sorting.
Note: To read data from KDDart the user must first be authenticated (i.e. logged in to the system). They must also have permission to read the data they are trying to retrieve.
In the example above, where all specimen names were being retrieved, the list returned would only contain those the user was permitted to read.
For example, KDDart may have 20,000 specimen names and the user’s ‘list all’ may only display 100 records, which are those they have permission to view).
Write (Add and Update) Operations
The DAL caters for KDDart write operations using the ‘add’ and ‘update’ operations, which are protected by data integrity security. This feature employs mechanisms using write tokens, granted by the Data Access Layer at login.
Put simply, the DAL uses the HMAC SHA1 algorithm for generating data security signatures and detects if the data entered on the client/user end matches what is received at the server before adding or updating to KDDart.
The following examples have been kept simple, without delving into the data structures containing the parameters and data for adding or updating. However, the URL syntax is simple as shown below.
There are array structures and POST parameters, not shown, which contain the data for add or update to KDDart.
The Add example shown next adds a new genotype, whereas the Update is updating a ‘trial’ record with trial id=1.
Add:
add/genotype
Update:
update/trial/1
String genotypeName = "DemoGenotype" + mDal.getRandomNumberString();
String genotypeId = postReturnVal;
String speciesName = "Demonstration Example";
String acronym = "none";
String originId = "0";
String canPublish = "0";
String genoNote = "Demonstration of adding a new genotype in Java";
String genoColor = "N/A";
String ownPerm = "7";
String accessGrpId = "0";
String accessPerm = "5";
String otherPerm = "5";
postReturnVal = mDal.add_record("/add/genotype", addGenotypeParameters);
This code representation is in a simplified form for presentation (especially setting static values), however it illustrates the common method of DAL interaction.
