DAL examples

The examples provided illustrate the method of accessing KKDart through the DAL API and are representative of the consistent and large range of published operations.
Table of Contents
Introduction
Objective
If you are preparing to use the Data Access Layer (DAL) Application Programming Interface (API) the Getting Started page is recommended reading to understand the concepts and functionality provided in the DAL API before commencing with the examples.
The examples provide a simple illustration of the method to access KKDart through the DAL API and are representative of the consistent and large range of published operations.
Audience
This document is only intended for Programmers/IT Developers needing to use KDDart to extend their existing programs/system functionality and/or to develop new applications as requirements dictate.
Using DAL
Preliminaries
Programmers need to be aware of the following DAL characteristics:
- DAL, as a web service interface to KDDart, is accessible via HTTPS or HTTP depending on the chosen configuration of your installation;
- The http client must be able to read/write cookies;
- The client must be able to generate a Hash-based Message Authentication Code (HMAC) using SHA1 hashing algorithm;
- DAL returns the results of calls as XML inside
<DATA>tags.; and - DAL URLs follow the REST convention (hence RESTful API). For example the URL construction to list the fields in the specimen table would be similar to:
https://KDDart.YourURL.com/dal/specimen/list/field
What You Need To Know
To make a quick start to commence programming with DAL the following guide is provided to assist.
It is suggested that you work through the topics in the following table in the order presented:
| Step | Topic | Description |
|---|---|---|
| 1 | Requirements for the Example | Provides a brief scenario of what the example is to perform |
| 2 | Status and Error Handling | Describes how errors are handled and results and conditions are returned to your program |
| 3 | DAL Authentication | How to authenticate/login with DAL (Group Admin access is required) |
| 4 | Set Group | Using Switch Group to set the Group following login |
| 5 | DALFuncAddGenotype | Adding a genotype to the database table and checking the returned result |
| 6 | DALFuncListGenotype | List the Genotypes. Confirm the success of the previous 'add genotype' |
| 7 | DALOpLogout | Logout or disconnect from DAL |
The programming examples illustrate a series of common DAL functions.
These include scenarios for success and 'intended to fail' to demonstrate condition and return information handling.
Before getting underway the error/results handling is covered first as this is required from the start and with all API calls in your program(s).
Requirements for the Example
Construct a sample program to illustrate DAL usage which will utilise the following API functions:
- Login to KDDart;
- Set the group for the session;
- Add (Genus and Genotype);
- List Genotype;
- Import Genotype; and
- Logout from KDDart.
Perform the following process flow:
| Step | Action | Description |
|---|---|---|
| 1 | Login | Login to DAL - Illustrating checking error/result conditions |
| 1a | Login Fail | Login with non existing user - Login fails |
| 1b | Login Fail | Login with wrong password - Login fails |
| 1c | Login Success | Login successfully |
| 2 | Set Group | Set the required Group following Login |
| 2a | Set Group fails | Set Group with non existing group |
| 2b | Set Group Success | Set Group with a 'valid' group |
| 3 | Add Data | Add Genus and Genotype - Illustrating checking error/result conditions |
| 3a | Add Genus succeeds | Successful add genus - No error |
| 3b | Add Genus fails | Add genus fails as genus already existing |
| 3c | Add Genotype Success | Genotype success |
| 4 | Read data | Get data |
| 4a | Get Genotype | Get the newly added genotype - Valid read |
| 4b | List Genotypes | List the last 5 genotypes - Valid read |
| 4c | Import Genotypes | Import genotype from a .csv file with 5 rows - Valid import |
| 4d | Update Genotype | Valid Update |
| 4e | Export Genotype | Valid Export |
| 4f | Delete Genotype | Valid Delete |
| 5 | Logout | Logout of DAL |
Status and Error Handling
This topic describes status codes and provides some Example Error Handling Code.
DAL error and return codes (ie status codes) or messages have the following characteristics:
- DAL error and return codes or messages will be returned via HTTP; and
- All messages are returned in a single XML element.
DAL Status and Error Handling is separated into the following categories:
| # | Category | Code | Description |
|---|---|---|---|
| 1 | Success | 200 | The status code 200 is the HTTP status code for success (see: https://tools.ietf.org/html/rfc2616#section-10.2) |
| 2 | DAL Authentication Error | 401 | These errors relate to authentication or permissions 'Errors' or returned messages intercepted and interpreted by DAL and returned as an XML message. The returned message should be similar to the following: <DATA><Error Message="Signature verification failed." /></DATA> |
| 3 | Unknown URLs | 404 | Caused by either:
|
| 4 | DAL Application Error | 420 | This range of errors are specific to the logic of the application and either dealt with by the program or the user. The 'Errors' or returned messages intercepted and interpreted by DAL and returned as an XML message as in the above example. |
| 5 | Apache Server Errors | 500 | Caused by either:
|
DAL will always return a result/message in XML or other format if requested.
- When no error occurs DAL returns a code 200 with data wrapped in an XML element. This element name is dependant on the name of the associated table;
- DAL also needs to return information acknowledging a successful request. This could be returned in an XML message like:
<Info Message="Genotype (157) has been added successfully." />
DAL 401 error relates to authentication errors and will occur when:
- A user name and/or password are non-existent/incorrect;
- After successful login a group is not selected and set then a DAL operation is attempted;
- When a DAL operation is performed that requires group administrator privilege and the current group does not have that access.
Note: DAL 420 result/errors are returned when DAL does not return a 200 or 401. The detail is in the returned XML message. It will depend upon what the program is attempting to do with DAL at the time and up to the logic of the program how the condition is dealt with.
Example Error Handling Code
The following sample code illustrates interrogation of the returned Statusline. Specific to Login, it illustrates error codes in the 401 range as well as in the 420 range.
The example shows:
- Status code 200 indicates success;
- Status code 401 is returned from DAL with a response appropriate to the request;
- Status code 420 is the coverall for returned conditions that logic should be prepared to deal with and may not actually be an 'error'; and
- Otherwise the native Statusline is returned (these messages cannot be managed by the DAL).
StatusLine statusLine = response.getStatusLine();
int errCode = statusLine.getStatusCode();
System.out.println("Status Code: " + errCode);
/**
* Check http status code for success or failure
*/
if (errCode == 200) {
/**
* Http status code 200: Login is successful
*/
Element elem = (Element) doc.getElementsByTagName("WriteToken").item(0);
if (elem != null)
{
writeToken = elem.getAttribute("Value"); //Set instance variable 'writeToken'
returnValue = writeToken;
}
}
else if (errCode == 401) {
/**
* Http status code 401: Authentication is unsuccessful, group is not assigned or the group administration privilege is missing for the current user/group
*/
Element elem = (Element) doc.getElementsByTagName("Error").item(0);
if (elem != null) {
String dalErrMsg = elem.getAttribute("Message");
System.out.println("DAL Error Message: " + dalErrMsg);
}
returnValue = "false";
}
else if (errCode == 420) {
/**
* Http status code 420: Returned condition result/error for checking by program
*/
Element elem = (Element) doc.getElementsByTagName("Error").item(0);
if (elem != null) {
String dalErrMsg = elem.getAttribute("Message");
System.out.println("DAL Error Message: " + dalErrMsg);
}
returnValue = "false";
}
else {
System.out.println("Status Line: " + statusLine.toString());
returnValue = "false";
}
DAL Authentication
This section describes how authentication works in DAL, along with how to construct Login and Set Group code. It is important to understand both for login and also regarding the write token which is needed to change/add data to KDDart.
The following Permission Matrix shows task that can be performed for different user types:
| Task | Admin and a Manager | Admin and NOT a Manager | Manager | User | Guest |
|---|---|---|---|---|---|
| See all records regardless of the record permission | Yes | Yes | No | No | No |
| Change record permission regardless of the permission | Yes | No | No | No | No |
| Add and remove users, add and remove groups, add and remove users from a group and reset user password | Yes | No | No | No | No |
| See their own records | Yes | Yes | Yes | Yes | No |
| Update their own records | Yes | Yes | Yes | Yes | No |
| Change permission of their own records | Yes | No | Yes | No | No |
| Add and update types, design, breeding method etc. (vocabulary entities) | Yes | No | Yes | No | No |
| See public records | Yes | Yes | Yes | Yes | Yes |
Authentication Construction
DAL Authentication has the following characteristics:
- Hash-based Message Authentication Code (HMAC) with SHA1 hashing algorithm is used;
- User passwords are NOT stored on the system
- User passwords are NOT sent to the server
- KDDart uses authentication and permissions to provide record level access controls within the database.
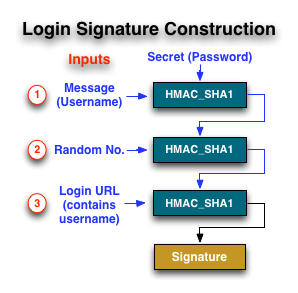
The following diagram illustrates the construction of a signature.
Signature construction only occurs on the 'client side' and at NO TIME is the user's password sent to the server.
Login to KDDart Using DAL
This section describes how to login to KDDart using DAL to establish an active session.
The following table describes the steps required to login using DAL:
| Step | Action | Description |
|---|---|---|
| 1 | Derive signature (a) | Calculate HMAC_SHA1 using the username as the data and the user's plain password as the key |
| 2 | Derive signature (b) | Calculate HMAC_SHA1 using the value in “rand_num” as the data and the result from step (a) as the key Note: rand_num: can be any number |
| 3 | Derive signature (c) | Calculate HMAC_SHA1 using the url as the data and the result from step (b) as the key url: according to the login URL syntax. |
| 4 | Setting POST parameters | POST requires the following three parameters to be set:
|
| 5 | Submit Login to server | Login has the following syntax: |
| 6 | Check Error Code | Check for successful Login |
| 7 | Save the 'Write Token' | Once the authentication is completed successfully, DAL issues several cookies and a 'write token' in the reply message. The client must save this 'write token' in a secure location for later use for KDDart updates. The 'write token' for the same session is required to sign data before being passed to the server for updating KDDart (i.e. performing Add, Update, Delete or Import operations). |
Note: HMAC_SHA1 must produce a hexadecimal string rather than binary data in order for the signature to be matched with the server.
The result would be similar to 90b48e51863dfcdb1b8125f02b9b8e270c3d845c.
Note: The hex string must be in lower case.
<?xml version="1.0" encoding="UTF-8"?> <DATA><WriteToken Value="7fd825c99c8abeac9dc4d9b2aea4490f1578cb36" /> <User UserId="0" /></DATA>
Examples
The following example Java code illustrates the Login operation:/*
* The user is logged into KDDart.
*/
int LoginErr = mAuth.Login(mHttpClient, dalBaseUrl);
if (LoginErr != 1) {
System.out.println("Login is successful");
}
Note: If the username or the password is rejected, DAL returns an XML error message: “Incorrect username or password.”. Refer to Status and Error Handling for further details.
Status/error results from the List operation are as follows:
package org.dart.kddart.dal;
import java.io.IOException;
import javax.xml.parsers.ParserConfigurationException;
import org.apache.http.client.HttpClient;
import org.dart.kddart.dal.auth.Auth;
import org.xml.sax.SAXException;
public class Login
{
/**
* Tries to login
* @param baseUrl The base url of the service
* @param username The username to use
* @param password The password to use
* @return true of the login was successful
* @throws SAXException
* @throws ParserConfigurationException
* @throws IOException
* @throws IllegalStateException
*/
public static boolean login(String baseUrl, String username, String password)
throws IllegalStateException, IOException,
ParserConfigurationException, SAXException
{
HttpClient mHttpClient = KDDArTHTTPClient.getInstance();
Auth auth = Auth.getInstance();
return auth.login(mHttpClient, baseUrl, username, password);
}
}
Set Group
Groups define access privileges in KDDart - what the user can view or add/change.
Users may belong to multiple groups so after login the user must set the group to use.
When setting the group an error will be returned if the:
- User is not a member of the group being set; or
- Group supplied does not exist.
The following code sample illustrates the setting of a group using DAL:
/**
* Sets the group for the user
*
* @param hClient
* The client used for the connection
* @param baseUrl
* The base url
* @param groupNum
* The group number
* @return The log
* @throws IOException
* @throws ClientProtocolException
*/
public String setGroup(HttpClient hClient, String baseUrl, int groupNum)
throws ClientProtocolException, IOException
{
String url = baseUrl + "/switch/group/" + String.valueOf(groupNum);
HttpPost post = new HttpPost(url);
HttpResponse response;
response = hClient.execute(post);
BufferedReader reader = new BufferedReader(new InputStreamReader
(response.getEntity().getContent()));
String output = "";
String line;
while ((line = reader.readLine()) != null)
{
output += line;
}
return output;
}
Add Genotype
Description
This example illustrates the:
- Addition of a new Genotype record to the Genotype table;
- Preparation of the data parameters (attribute construction consists of building a name value pair array); and
- Uses the generic DAL function mDal.add_record to record the data in the Genotype table.
Prerequisites
In preparing software to add a record to KDDart:
- The user must be authenticated (i.e. Logged in to KDDart);
- A 'write token', specific to the 'current' session which is issued at login, must have been saved to allow generation of a signature for the Add and also Update operations;
- The user must have the appropriate Group set to allow the record to be added. Refer to for further information regarding permissions;
- Knowledge of all the column details to be added to the table (can be determined by using the list/field operation).
Example Code - Parameter Preparation
This example shows preparation of a name value pair array to add a new Genotype record:
String genotypeName = "GenoTest " + mDal.getRandomNumberString();
String genusId = postReturnVal;
String speciesName = "Testing";
String acronym = "none";
String originId = "0";
String canPublish = "0";
String genoNote = "Testing from Java";
String genoColor = "N/A";
String ownPerm = "7";
String accessGrpId = "0";
String accessPerm = "5";
String otherPerm = "5";
List<NameValuePair> addGenotypeParameters;
addGenotypeParameters = new ArrayList<NameValuePair>(1);
addGenotypeParameters.add(new BasicNameValuePair("GenotypeName", genotypeName));
addGenotypeParameters.add(new BasicNameValuePair("GenusId", genusId));
addGenotypeParameters.add(new BasicNameValuePair("SpeciesName", speciesName));
addGenotypeParameters.add(new BasicNameValuePair("GenotypeAcronym", acronym));
addGenotypeParameters.add(new BasicNameValuePair("OriginId", originId));
addGenotypeParameters.add(new BasicNameValuePair("CanPublishGenotype", canPublish));
addGenotypeParameters.add(new BasicNameValuePair("GenotypeNote", genoNote));
addGenotypeParameters.add(new BasicNameValuePair("GenotypeColor", genoColor));
addGenotypeParameters.add(new BasicNameValuePair("OwnGroupPerm", ownPerm));
addGenotypeParameters.add(new BasicNameValuePair("AccessGroupId", accessGrpId));
addGenotypeParameters.add(new BasicNameValuePair("AccessGroupPerm", accessPerm));
addGenotypeParameters.add(new BasicNameValuePair("OtherPerm", otherPerm));
postReturnVal = mDal.add_record("/add/genotype", addGenotypeParameters);
if (postReturnVal != "failed") {
System.out.println("New genotype Id for " + genotypeName + " is: " + postReturnVal);
}
List Genotype
Description
Managing KDDart information using DAL helps simplify programming effort through the use of generic routines. This example illustrates reading KDDart information from the Genotype table, however the same construction is used for other tables.
DAL has two functions for retrieving data from KDDart:
- List to retrieve multiple records
list/<table name>[/<number of records per page>/page/<page number>] - Get to retrieve a single specific record
get/<table name>/<record id>
The following example:
- Illustrates the Java code for listing Genotypes;
- is an extract from the example code; and
- Shows the resultant output captured from executing the code sample.
The first output result is singular, from listing a specific Genotype. The second output result is from listing Genotypes with a limit of five.
Example Code
Example Step 4
/*
* Example Step 4
*
* This step has the following sub-steps
* -a: Get the newly added genotype
* -b: List the last 5 genotypes
*/
System.out.println("4a - Starts");
String newGenotypeId = postReturnVal;
String getReturnVal = mDal.read("/get/genotype/" + newGenotypeId);
if (getReturnVal != "failed") {
System.out.println("List result:\n" + getReturnVal);
}
System.out.println("4a - Finishes");
System.out.println("4b - Starts");
getReturnVal = mDal.get("/list/genotype/5/page/1");
if (getReturnVal != "failed") {
System.out.println("List result:\n" + getReturnVal);
}
System.out.println("4b - Finishes");
Example Code Output 4a
Note: The following text has been 'massaged' with line breaks for visual clarity.
Example XML Code Output Labelled 4a
4a - Starts Status Code: 200 List result: <?xml version="1.0" encoding="UTF-8"?> <DATA> <RecordMeta TagName="Genotype" /> <Genotype AccessGroupPerm="5" AccessGroupId="0" GenotypeId="1587" GenotypeName="GenoTest -1268469055" AccessGroupPermission="Read/Link" OtherPermission="Read/Link" addAlias="genotype/1587/add/alias" chgPerm="genotype/1587/change/permission" OtherPerm="5" OwnGroupPerm="7" CanPublishGenotype="0" OriginId="0" SpeciesName="Testing" GenotypeNote="Testing from Java" GenotypeColor="N/A" OwnGroupPermission="Read/Write/Link" OwnGroupName="admin" GenusName="GenusTest2 - 598721270" GenusId="47" AccessGroupName="admin" GenotypeAcronym="none" chgOwner="genotype/1587/change/owner" UltimatePermission="Read/Write/Link" delete="delete/genotype/1587" OwnGroupId="0" update="update/genotype/1587" UltimatePerm="7" /> </DATA> 4a - Finishes
Example Code Output 4b
Note: The following text has been 'massaged' with line breaks for visual clarity.
Example XML Code Output Labelled 4b
4b - Starts Status Code: 200 List result: <?xml version="1.0" encoding="UTF-8"?> <DATA> <Pagination Page="1" NumOfRecords="1587" NumOfPages="318" NumPerPage="5" /> <RecordMeta TagName="Genotype" /> <Genotype AccessGroupPerm="5" AccessGroupId="0" GenotypeName="GenoTest -1268469055" GenotypeId="1587" AccessGroupPermission="Read/Link" OtherPermission="Read/Link" addAlias="genotype/1587/add/alias" chgPerm="genotype/1587/change/permission" OwnGroupPerm="7" OtherPerm="5" CanPublishGenotype="0" OriginId="0" GenotypeNote="Testing from Java" SpeciesName="Testing" GenotypeColor="N/A" OwnGroupPermission="Read/Write/Link" OwnGroupName="admin" GenusName="GenusTest2 - 598721270" GenusId="47" AccessGroupName="admin" GenotypeAcronym="none" chgOwner="genotype/1587/change/owner" UltimatePermission="Read/Write/Link" delete="delete/genotype/1587" OwnGroupId="0" update="update/genotype/1587" UltimatePerm="7" /> <Genotype AccessGroupPerm="5" AccessGroupId="0" GenotypeName="GenoTest -2045377762" GenotypeId="1586" AccessGroupPermission="Read/Link" OtherPermission="Read/Link" addAlias="genotype/1586/add/alias" chgPerm="genotype/1586/change/permission" OwnGroupPerm="7" OtherPerm="5" CanPublishGenotype="0" OriginId="0" GenotypeNote="Testing from Java" SpeciesName="Testing" GenotypeColor="N/A" OwnGroupPermission="Read/Write/Link" OwnGroupName="admin" GenusName="GenusTest2 - 351388315" GenusId="45" AccessGroupName="admin" GenotypeAcronym="none" chgOwner="genotype/1586/change/owner" UltimatePermission="Read/Write/Link" delete="delete/genotype/1586" OwnGroupId="0" update="update/genotype/1586" UltimatePerm="7" /> <Genotype AccessGroupPerm="5" AccessGroupId="0" GenotypeName="GenoTest 608111755" GenotypeId="1585" AccessGroupPermission="Read/Link" OtherPermission="Read/Link" addAlias="genotype/1585/add/alias" chgPerm="genotype/1585/change/permission" OwnGroupPerm="7" OtherPerm="5" CanPublishGenotype="0" OriginId="0" GenotypeNote="Testing from Java" SpeciesName="Testing" GenotypeColor="N/A" OwnGroupPermission="Read/Write/Link" OwnGroupName="admin" GenusName="GenusTest2 - -315807079" GenusId="43" AccessGroupName="admin" GenotypeAcronym="none" chgOwner="genotype/1585/change/owner" UltimatePermission="Read/Write/Link" delete="delete/genotype/1585" OwnGroupId="0" update="update/genotype/1585" UltimatePerm="7" /> <Genotype AccessGroupPerm="5" AccessGroupId="0" GenotypeName="GenoTest 1106528240" GenotypeId="1584" AccessGroupPermission="Read/Link" OtherPermission="Read/Link" addAlias="genotype/1584/add/alias" chgPerm="genotype/1584/change/permission" OwnGroupPerm="7" OtherPerm="5" CanPublishGenotype="0" OriginId="0" GenotypeNote="Testing from Java" SpeciesName="Testing" GenotypeColor="N/A" OwnGroupPermission="Read/Write/Link" OwnGroupName="admin" GenusName="GenusTest2 - -2033749030" GenusId="41" AccessGroupName="admin" GenotypeAcronym="none" chgOwner="genotype/1584/change/owner" UltimatePermission="Read/Write/Link" delete="delete/genotype/1584" OwnGroupId="0" update="update/genotype/1584" UltimatePerm="7" /> <Genotype AccessGroupPerm="5" AccessGroupId="0" GenotypeName="GenoTest 764902838" GenotypeId="1583" AccessGroupPermission="Read/Link" OtherPermission="Read/Link" addAlias="genotype/1583/add/alias" chgPerm="genotype/1583/change/permission" OwnGroupPerm="7" OtherPerm="5" CanPublishGenotype="0" OriginId="0" GenotypeNote="Testing from Java" SpeciesName="Testing" GenotypeColor="N/A" OwnGroupPermission="Read/Write/Link" OwnGroupName="admin" GenusName="GenusTest2 - 1812372840" GenusId="39" AccessGroupName="admin" GenotypeAcronym="none" chgOwner="genotype/1583/change/owner" UltimatePermission="Read/Write/Link" delete="delete/genotype/1583" OwnGroupId="0" update="update/genotype/1583" UltimatePerm="7" /> </DATA> 4b - Finishes
More code examples
To see more examples and applications, please go to the Downloads page.
